|
||
      |
Gene counter
Geneticist Jonathan Pritchard tallies the evidence for recent human evolution.
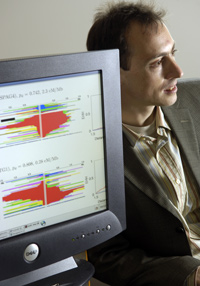
Using statistical analysis, Jonathan Pritchard identified 700 “signals
of selection” that show evolutionary changes in the human genome
in the last 6,000 to 10,000 years.
Jonathan Pritchard is philosophical—and perhaps a little puzzled—about the media attention surrounding his announcement that the human genome has continued to adapt over the last 6,000 to 10,000 years, the blink of an eye in evolutionary time. “Still evolving, human genes tell new story,” proclaimed a March 7 New York Times headline; on July 24 the Washington Post declared, “And the evolutionary beat goes on...” For Pritchard, the idea that evolution might persist today comes as no surprise. The human-genetics professor and his research team found more than 700 signals of recent natural selection—mathematical indicators that mutations were on their way to becoming permanent—in areas relating to reproduction, brain development, skin pigmentation, and food metabolism. “One thing newspapers were interested in,” he says, “was this idea that humans have been evolving within recent history, which is something I sort of presumed.”
Published this past March in Public Library of Science-Biology (PLoS–Biology), Pritchard’s report provides a “first-generation map,” he says, of “signals of selection across the human genome.” The paper also explores which genes fall within those signals, what kinds of biological processes those genes are involved in, and how the mutations might connect to evolutionary pressures.
For 99.9 percent of any human chromosome, DNA base-pair sequences are identical. But 0.1 percent vary from person to person; many of those variations are known as single nucleotide polymorphisms or SNPs (pronounced “snips”). By 2002 researchers had identified about 10 million common SNPs, “fueling studies of human variation,” says Pritchard, who earned a biology PhD from Stanford in 1998. For his PLoS–Biology report, he used data from the International HapMap Project, which collected DNA from several hundred people in four populations: Han Chinese from Beijing, Japanese from Tokyo, Yorubans from Nigeria, and Utah families with European ancestors. Because nearby SNPs on the same chromosome generally are inherited as a unit, HapMap researchers mapped these closely-related patterns of SNPs, or “haplotype blocks.”
Pritchard searched HapMap data for evidence of mutations selected within the last 10,000 years. A mutation, or alteration in DNA, happens initially in one individual. If it has no impact on survival, it spreads very slowly or—most likely—disappears. But if it aids survival, Pritchard says, there is a “relatively short time from when one individual has it to when everyone in the population has it.” Recent useful mutations, he reasons, are in the process of spreading rapidly but have not yet become fixed within a population.
How to find those positive mutations? Pritchard, along with lab members Ben Voigt and Sridhar Kudaravalli, developed a statistical-analysis method based on predictable variation patterns in the SNPs around a chromosome’s mutation. As it spreads through a population, a mutation carries with it the surrounding haplotype block. If the beneficial mutation occurred a long time ago, other parts of this haplotype will have become “jumbled up” as a result of normal recombination/mutation, says Pritchard. But if the mutation is relatively new, the haplotype will look the same from person to person, since it won’t have had a chance to develop other changes. What he calls a “signal of selection” is this statistically significant uniformity in a section of a chromosome’s genetic sequence.
Pritchard reports finding plenty of evidence that humans have continued to evolve in response to changing environments and cultures. Signals of selection show up in all HapMap population groups, although specific changed genes vary from one group to another. For instance, genes that enable adults to continue producing the enzyme lactase, which digests milk sugar, have been selected in Europeans, whose ancestors raised cattle and drank milk into adulthood. He also found five genes involved in skin pigmentation that show genetic changes in Europeans, “presumably reflecting when modern humans spread into Northern Europe” and began producing the pale skin needed to absorb sunlight for vitamin D synthesis. Genes connected to brain development and reproduction show changes in all population groups, although the specific genes differ.
Pritchard says his study raises more questions than it answers. “In many cases, we don’t know what the biological function [of a changed gene] is; in many cases we don’t know what the actual selective pressure is.” To make it easier to determine the functions of evolving genes, he’s preparing to analyze DNA samples from about ten different African populations. Africans have more genetic variation than non-Africans, Pritchard notes, and their signals of selection tend to be clearer, stronger, and narrower. The results, he hopes, will be easier to interpret.
The African-population study is one of several projects on tap for Pritchard’s lab. Using DNA data extracted from Neanderthal fossils, he’s trying to figure out whether Neanderthals and ancestors of modern Europeans ever mated (no definitive answer yet, but he says it’s “great fun” to investigate). Collaborating with Chicago geneticist Carole Ober, he’s also examining “copy number variation,” a genetic deviation in which DNA segments are either missing or repeated in some individuals. Their goal is to answer a basic question: how do copy number variations relate to disease?
New kinds and quantities of data “we couldn’t have imagined collecting two years ago” continue to expand his research possibilities, says Pritchard. “Our real interest is to push the envelope to new ways of thinking about data, about how we can use these data to answer interesting biological questions.”